Errors in genotype scoring
 (after Stam, Study guide of the Wageningen University MSc course Modern statistics for the life sciences)
(after Stam, Study guide of the Wageningen University MSc course Modern statistics for the life sciences)
Errors in genotype scoring, or genotype classification, will lead to incorrect estimates of recombination frequencies, and hence, incorrect genetic distances. We illustrate this effect by means of a simple numerical example. Suppose the following situation:
- We want to determine the genetic distance between a locus for flower color (blue A/ red a) and seed coat (wrinkled B / smooth b). We make a backcross between a double heteozygous blue, wrinkled plant to a red, smooth plant: AaBb x aabb
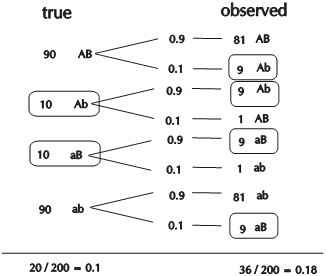
- The true recombination frequency between these loci is 10% (r = 0.10) In the figure on the right, recombinant gametes are boxed.
- Let us assume that the difference between wrinkled and smooth (B/b) is not so easy to observe and that mistakes are made in 10% of the cases:
- Of 90 AB plants, which are in fact blue and wrinkled (left in the figure), 81 are correctly scored blue and wrinkled and 9 are erroneously scored blue and smooth (right in the figure).
- Of 10 Ab plants, which are in fact blue and smooth, 9 are correctly scored blue and smooth and 1 is erroneously scored blue and wrinkled.
- Etc.
In reality, there would be 20 recombinants (blue+smooth and red+wrinkled) out of 200 pollen grains (figure, left). From the score, it appears that 36 out of 200 would be recombinants
(figure, right). So, in case of 10% erroneous scoring, the estimate of r (0.18) is almost twice as large as the real value! It should be noted that when recombinants are incorrectly scored as non-recombinants, the recombination frequency estimate would actually decrease instead of increase. However, as in the example, if the error scoring is independent of the true genotype, and there are fewer recombinants than non-recombinants (by definition), the non-recombinants would be affected more than the recombinants: in this example, from the 180 true non-recombinants, 18 are now counted as recombinants. From the 20 true recombinants, 2 are now counted as non-recombinants, so this results in a net balance of 16 extra recombinants on a total of 200 individuals.
Similar deviations in calculated distances occur when marker alleles are erroneously scored.
Show/hide comprehension question...
Summary
→ Errors in genotype scoring result in relatively large errors in estimates of recombination frequencies (= overestimations) and map distances