Alternative explanation: linkage analysis in practice
Markers on non-homologous chromosomes inherit independently, because of independent assortment of chromosomes. Markers on homologous chromosomes inherit in association with each other (unless they are located at a large distance on that chromosome: in that case, for a diploid the probability of recombination tends to 0.5, corresponding to independent inheritance and equal to the situation for two different chromosomes). Markers on homologous chromosomes can be exchanged through crossover (during meiosis). The larger the distance between two markers, the higher the probability of a crossover occurring between them. If one crossover occurs between two markers, the alleles of the parents recombine.
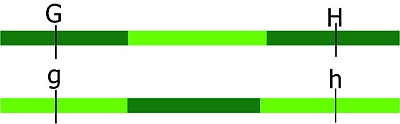
Imagine two genes/markers, G and H, which are both located on the same chromosome. Imagine an individual resulting from a cross between Parent 1 (GGHH) and Parent 2 (gghh). This individual will have received GH from Parent 1 and gh from Parent 2 and thus wil be GgHh. The gametes formed by this individual will be as follows:
NO CROSS-OVER results in gametes GH and gh (both scored as 'parental'), expected to occur in equal proportions.
ONE CROSS-OVER between G/g and H/h results in gametes Gh and gH (both scored as 'recombinant') in equal expected proportions.
TWO CROSS-OVERS between G/g and H/h results in GH and gh gametes. These are also scored as 'parental' or non-recombinant, because only the markers are scored and nothing is observed about the piece of DNA in between the markers.
|
|
Two gametes produced after double cross-over. Generally: odd numbers of cross-overs result in recombinant genotypes, even numbers in non-recombinant genotypes. The occurrence of more than three cross-overs per arm is rare, even for large chromosomes. |
Map distances are estimated by computer programs based on recombination frequencies, estimated from observed genotype frequencies (fractions of individuals that have a certain genotype for a combination of two markers). The larger the true distance between two genes (or markers), the less precise the estimated distance will be, because higher recombination frequencies have higher sampling variance.