Genetic map compared to physical map
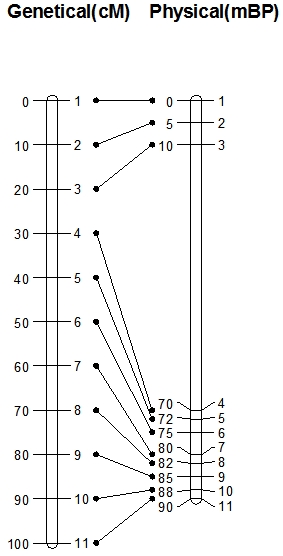 In the picture on the left you see two types of maps: on the left hand side you see a genetic map with 11 evenly spaced markers (1-11). The space between the markers is measured in centiMorgans, which, at short distances (up to about 10 cM) approximately represent the percentage of recombination. The distance between, for example, Marker 1 and Marker 2 is 10 cM, so the percentage of recombination between Marker 1 and Marker 2 is about 10%. Marker 1 and Marker 2 are therefore linked and the large majority of gametes carry the parental combinations of alleles. However, this does not automatically mean that Marker 1 and 2 lie physically closely together on the chromosome! This is visualized in the physical map of the same chromosome on the right hand side. Physical maps are based on the distances expressed in number of base pairs of the DNA. On the physical map in the picture, you see again the positions of Markers 1-11 and their positions, measured in millions of base pairs counted from the top. Marker 1 and Marker 2 are 5 million base pairs apart.
In the picture on the left you see two types of maps: on the left hand side you see a genetic map with 11 evenly spaced markers (1-11). The space between the markers is measured in centiMorgans, which, at short distances (up to about 10 cM) approximately represent the percentage of recombination. The distance between, for example, Marker 1 and Marker 2 is 10 cM, so the percentage of recombination between Marker 1 and Marker 2 is about 10%. Marker 1 and Marker 2 are therefore linked and the large majority of gametes carry the parental combinations of alleles. However, this does not automatically mean that Marker 1 and 2 lie physically closely together on the chromosome! This is visualized in the physical map of the same chromosome on the right hand side. Physical maps are based on the distances expressed in number of base pairs of the DNA. On the physical map in the picture, you see again the positions of Markers 1-11 and their positions, measured in millions of base pairs counted from the top. Marker 1 and Marker 2 are 5 million base pairs apart.
On the genetic map, Markers 2 and 3 are 10 cM apart, and Markers 3 and 4 also. However, on the physical map, we see that the genetic distance does not predict at all the physical distance. Distance between Marker 2 and 3 is about 5 million base pairs, and distance between Marker 3 and 4 is 60 million base pairs. This implies that in the long stretch of DNA between Marker 3 and 4 as many recombinations occur as on the much shorter stretch between Marker 2 and 3. So, per million base pairs, recombination is much lower between markers 3 and 4 than between 2 and 3.
Genetic maps are generated using the pairwise recombination frequencies between markers. Physical maps are generated by sequencing: the order of the DNA base pairs is assessed by sequencing machines and put together. When the sequences of markers used in a genetic map are known, they can be located on the physical map, and their positions may be estimated or determined.
Recombination hot spots and cold spots
During meiosis, cross-overs can occur, resulting in recombination. As will be seen below, not every crossover will be observed in the progeny. The probabilities of cross-overs occurring are not the same along the chromosome, as illustrated above. Usually fewer cross-overs per million base pairs occur close to the centromere, compared to the regions further away from the centromere. If many cross-overs occur within a short stretch of DNA, this results in a long distance on the genetic map. This region is then called a recombination hot spot. If few cross-overs occur on a long stretch of DNA, this results in a small distance on the genetic map, whereas the physical distance is great. This region is called a recombination cold spot. This explains the large distance between markers 3 and 4 on the physical map compared with the relatively short distance on the genetic map.
Show/hide comprehension question...
Reasons for large gaps in genetic maps: non-polymorphic DNA regions
|
|
|
|
|
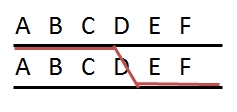
In non-polymorphic DNA regions, recombinations remain unnoticed (top). In the flanking polymorphic regions (A/a and I/i) we may find polymorphic markers to determine recombination frequencies. The (polymorphic) markers closest to the non-polymorphic region and flanking it (A/a and I/i), may recombine frequently, especially when the non-polymorphic region is relatively long. |
In the previous section, we saw that a recombination hot spot can result in relatively large distances in genetic maps. This is caused by many recombinations occurring in a relatively short stretch of DNA (physical map).
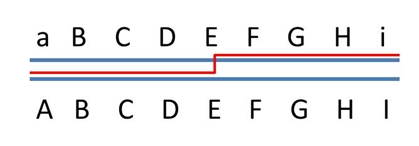
When both chromosomes of a diploid plant have an identical stretch of DNA, i.e. no polymorphism there, cross-over events may occur in that region, but this will remain unnoticed in the progeny, since in that region no markers can be developed (picture on the right: top). Still recombinations in that area occur, and will result in recombination between the polymorphic flanking areas (picture on the right: bottom) if there are any. Therefore the non-polymorphic area will result in a gap in the linkage map, since the flanking markers have a rather high percentage of recombination, and no markers occur in between them. If there are non-polymorphic regions at the ends of chromosomes, these will not be represented in the genetic map. Non-polymorphic regions may occur if two parents of the mapping population have one or more DNA fragments in common, because they have a common ancestor. Heterozygous regions are needed to observe recombination events in the resulting progeny.
Alternatively, a DNA fragment may have no homology with the sister chromosome. This may be the case in a introgression from a different, related, species. In such a region, there will be (almost) no chromosome pairing in the meiosis, and hence (almost) no recombination (picture below). This will result in a cold spot. Markers may be developed due to the high polymorphism, but they will cluster on the linkage map, since they are at distances of 0 cM from each other.
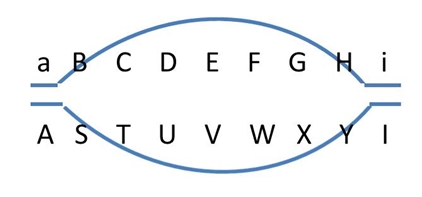
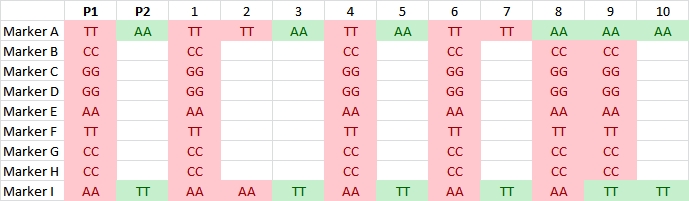
Illustration of two SNP markers segregating in a RIL or DH population (data of 10 individuals shown). They have no corresponding sequence in parent P2. The identical segregation pattern indicates that the two markers are located on the same DNA fragment that probably is not homologous between the two parents.
Summary
→ Genetic maps are expressed in centiMorgan units and are calculated from the pairwise recombination frequencies among markers and/or genes segregating in a progeny
→ Physical maps are expressed in base pair units and generated by sequencing
→ Recombination hot spots are regions on the chromosome (= on the physical map) where relatively many recombinations occur
→ In non-polymorphic DNA regions cross-overs occur, but their effect in terms of recombination may only be noticed in the polymorphic flanking regions of that segment or go unnoticed entirely if there are no polymorphisms
Show/hide comprehension question...
Show/hide comprehension question...