Development of a mapping population in autogamous species
In autogamous species, mapping populations usually originate from the F1 of a cross of inbred lines: each parent is homozygous and the F1 is heterozygous at the loci at which the parents have different alleles. The segregation among these parental alleles when the F1 pollen and F1 egg cells are formed is studied in a linkage analysis of genotypes in F1 derived populations, such as the ones shown below.
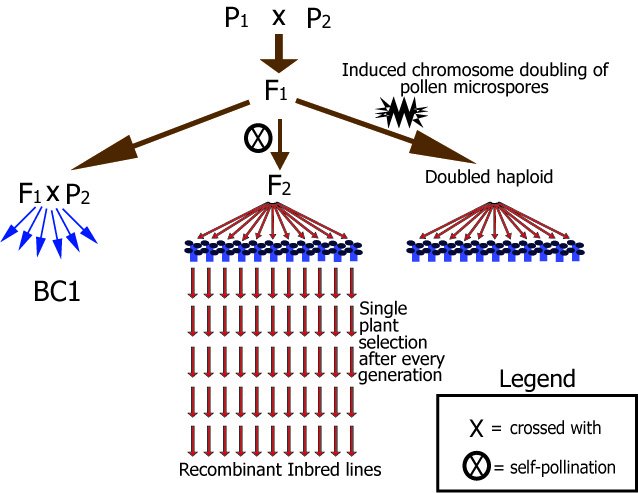
Diagram of main types of mapping populations for autogamous species.
In this figure, for example, Parent 1 (P1) could be AABBCCDDEEFFGGhh and Parent 2 aabbccddeeffGGhh, each letter representing an allele of a gene or marker. These examples are chosen for simplicity, but any other homozygous combination like AAbbccDDEEffGGhh and aaBBCCddeeFFGGhh could also be possible, as long as there is sufficient difference between the parents: in this example, polymorphism is present for six markers or genes (and absent for two). The resulting F1 population consists entirely of AaBbCcDdEeFfGGhh plants. Note that in this example the F1 is homozygous for the G and H locus only. After that, there are different ways to create segregating populations:
a. Selfing the F1 to create an F2
|
Population type |
Codominant markers |
Dominant markers |
|---|---|---|
|
F2 |
1 : 2 : 1 (AA:Aa:aa) |
3 : 1 (B_:bb) |
For a codominant gene or marker (for which AA plants can be distinguished from Aa plants), Aa x Aa results in an expected segregation of 1:2:1 (AA:Aa:aa).
For a dominant gene or marker Bb x Bb results, by expectation, in 3:1 (B_:bb).
F2 progenies in autogamous crops have been often used for construction of genetic maps and for studying monogenic traits. They are less useful for QTL analysis, genetic studies of quantitative traits or traits requiring much plant material of a , for two reasons:
- Each genotype is represented by only one plant, and each plant is genotypically unique. In most crops, single plants cannot be used for traits that need to be measured in several replications, such as yield and drought tolerance.
- Most crops are annual. So, each F2 plant will die at the end of the season: the population is "mortal". If we harvest seeds of each F2 plant, the progeny is not identical to each other, nor identical to the selfed plant, since selfing an F2 plant leads to more homozygosity and segregation.
F2 can be suitable for such analyses if the plant species is a perennial or if it can be vegetatively propagated to create clones. Sometimes the F2 is used for map construction while phenotyping is done in the F3 lines derived from genotypes F2 plants.
Selfing of individual F2 plants for a number of generations allows the development of recombinant inbred lines (RILs), which consist of a series of homozygous lines, which can be reproduced identically. It should be mentioned that some heterozygosity may still be present in RIL lines, especially if they are not from many generations of selfing.
How to develop RILs?
In a population of about 200 F2 plants, we harvest one seed per plant. Or better, two seeds to keep one as back-up, in case the first fails to germinate or to develop a next generation of seeds. We repeat this each generation. Harvesting and propagating in each generation one seed and one plant per original F2 plant is called single seed descent (see module on Principles of Plant Breeding - Selection methods; abbreviated to SSD).
At about the F7 or F8 we may presume that each of the plants is homozygous for the great majority of loci for which the parents originally carried different alleles. In the F7, a plant is probably heterozygous for about 1/64 of all loci that segregated in the F2.
The resulting set of about 200 homozygous plants, each derived from a different F2 plant of the same cross can be propagated by selfing. So, they can be phenotyped at population level in replications, and maintained infinitely: they are "immortal".
(Of course, an 'immortal' line can die, as a result of disasters or extreme weather conditions killing all plants, and when also the stored seed is lost). "Immortal" here refers to the genotype of the individuals, that can be reproduced identically for many generations, not to the individuals themselves, and as a contrast to the F2, for example, which can not be reproduced identically by fertilized seeds in the following generation.
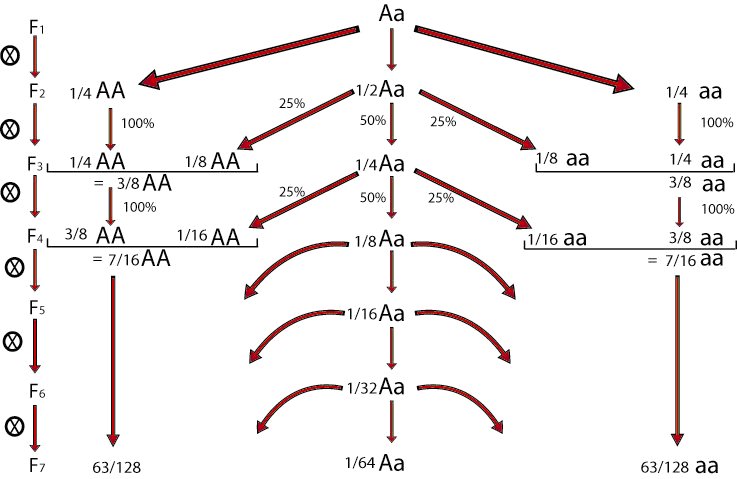
After six to eight generations, there will be homozygosity at most loci:
Plants homozygous at a locus produce 100% homozygous offspring for that locus and plants heterozygous at a locus produce 50% homozygous and 50% heterozygous offspring for that locus. The proportion of heterozygotes is halved in each generation. In every generation there is meiosis and there are possibilities of recombination as long as a chromosome segment still has heterozygous regions.
The resulting RILs each contain a unique combination of chromosomal segments from the original parents.
b. BC1: Backcrossing of the F1 to Parent 2
|
Population type |
Codominant markers |
Dominant markers |
|---|---|---|
|
F2 |
1 : 2 : 1 (AA:Aa:aa) |
3 : 1 (B_:bb) |
|
Backcross BC1 |
1 : 1 (Cc:cc) |
1 : 1 (Dd:dd) |
For a codominant gene or marker Cc x cc results in a segregation of 1:1 (Cc:cc).
For a dominant gene Dd x dd this is exactly the same: 1: 1 (Dd:dd) (Table).
Note that if the F1 is backcrossed to recurrent Parent 1 (Cc x CC), the result is a segregation of 1:1 (CC:Cc) for a co-dominant marker. For dominant markers we get 100% D_. So only markers can be used to genotype the mapping population that are co-dominant or for which the recurrent parent is homozygous recessive.
Note that the backcross population is compared to the F2 population equally mortal and equally confined to individual, genotypically unique plants.
c. Doubled haploids (DH)
|
Population type |
Codominant markers |
Dominant markers |
|---|---|---|
|
F2 |
1 : 2 : 1 (AA:Aa:aa) |
3 : 1 (B_:bb) |
|
Backcross |
1 : 1 (Cc:cc) |
1 : 1 (Dd:dd) |
|
Recombinant inbred lines or doubled haploids |
1 : 1 (EE:ee) |
1 : 1 (FF:ff) |
In a number of crop species, haploid plantlets are induced to develop from microspore cells or from certain prickle-pollinated egg cells. For several crops specific methods have been developed to induce such haploids out of male or female gametes. Those haploid plants may spontaneously or induced by chemicals double their chromosome numbers to result in a doubled haploid (DH). A segregating DH population can be obtained by inducing doubled haploids from an F1 plant (from a cross between homozygous parents). The Ee plants produce E and e gametes in roughly equal ratios. Doubling of chromosomes of the haploids results in 1:1 (EE:ee) plants, which segregate equally for the genes and markers for which the parents differed in allele. Note that this results in plants that are homozygous on all loci. Different plants however will be different homozygotes. For example, one plant might have the AAbbCCDDeeFFGGhh genotype whereas another might have the aabbCCddEEffGGhh genotype. The resulting set of about 200 DH plants are homozygous, each derived from a different gamete of the same heterozygous plant, and can be propagated identically by selfing. So, as with RILs they can be phenotyped at population level in replications, and maintained infinitely: they are "immortal".
Note that if F1 plants might be genetically different (because of non-homozygousity at some loci in a parent plant), the DH population should preferably be derived from a single F1 plant.
Advantages of RILs over DH
The DH plant is the result of only a single meiosis so that there is only one opportunity for recombination to occur: the meiosis in the F1 plant that produces the gametes that are used to induce the haploids. The next stage is already the DH population, in which selfing cannot increase the number of recombinations since the plants are already fully homozygous. There is a very different situation during the development of a RIL population. In a RIL population, there are not only the recombinations that occurred in the F1, but also in the heterozygous chromosome fragments in the F2 and in those in the F3 etc. The later the generation, the less chance for such heterozygous fragments in which additional recombinations may occur. Also, the plants at the start of the SSD procedure to create the RILs were the result of two gametes, and of the DH plant of only one gamete. The additional recombination opportunities together with the higher diversity at the start results in more recombinations than in the DH population. This results in a larger resolution of the map in RIL populations than in DH populations.
|
Population type |
Codominant markers |
Dominant markers |
|---|---|---|
|
F2 |
1 : 2 : 1 (AA:Aa:aa) |
3 : 1 (B_:bb) |
|
Backcross |
1 : 1 (Cc:cc) |
1 : 1 (Dd:dd) |
|
Recombinant inbred lines or doubled haploids |
1 : 1 (EE:ee) |
1 : 1 (FF:ff) |
d. Advantages and disadvantages of different types of mapping populations
→ Summary of advantages and disadvantages of mapping populations developed for autogamous species is listed in this table:
|
Mapping population |
Origin |
Advantages |
Disadvantages |
|---|---|---|---|
|
F2 |
F1 selfed |
|
|
|
Backcross (BC1) |
F1 x parent |
|
|
|
Recombinant inbred lines (RILs) |
Selfing of F2 for a number of generations |
|
|
|
Doubled haploid (DH) |
Induced chromosome doubling in gametes |
|
|