Linkage, recombination and genetic distance
During meiosis, genes and markers segregate via interchromosomal recombination (independent assortment of chromosomes) and intrachromosomal recombination (cross-over). Genes or markers that are on loci close to each other ('tightly linked') on the same chromosome will be transmitted together from parent to progeny more frequently than genes or markers that are located further apart on the same chromosome.
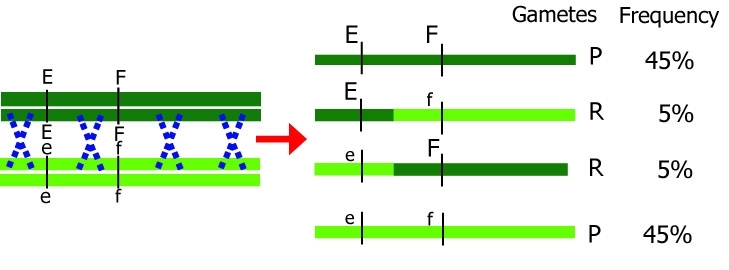
Effect of cross-over between homologous chromosomes.
Dark chromosomes originate from one parent; light chromosomes from the other parent. Only one chromosome pair is shown here. Note that during meiosis, each chromosome initially consists of two identical chromatids. P=parental, original, R=recombinant.
Gametes that are produced after meiosis either have the same combination as a parental chromosome (parental, P) or they have a different, non-parental combination (recombinant, R). The smaller the distance between two genes, the smaller the probability of recombination between the two genes.
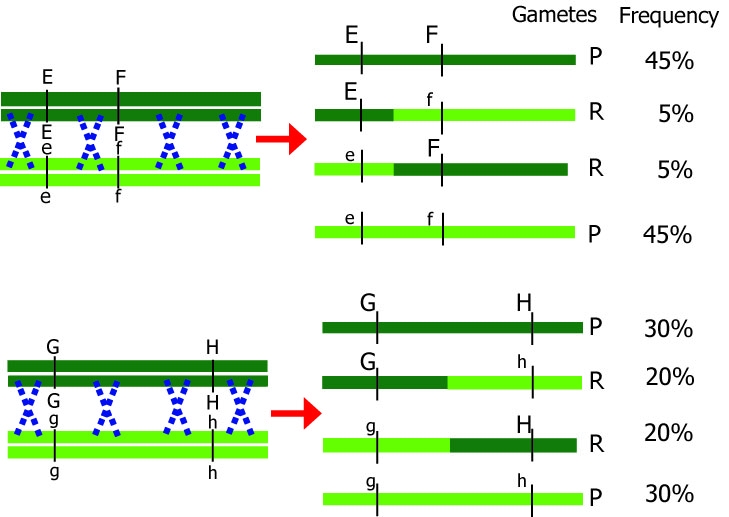
Different distances between genes result in different recombination percentages.
Note: The blue crosses in the figures do not indicate two recombinations at the same time, but rather that the chance for one recombination to occur is larger than when the loci of interest are closer to each other.
If loci are further apart, the chance of a crossover event occurring between them is larger. Therefore, recombination between loci G and H should occur more frequently than recombination between loci E and F. The other way around,
if we know the recombination frequencies, we convert these to genetic distances between the genes. Let us assume E, F, G, and H are loci in a population that segregates after crossing: from the number of recombinant individuals, it can be concluded that loci E and F are closer together than G and H.
More than one cross-over event can occur between loci. The probability of multiple recombinations increases with the distance between the loci. If two cross-over events take place between loci G and H in the same pair of chromatids, we would observe a non-recombinant genotype while in fact there was a double recombination event:
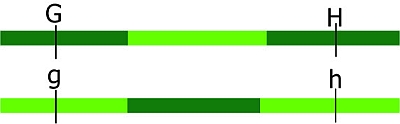
Keep in mind that: the closer to each other two loci are situated on a chromosome, the lower the recombination
frequency is. (And, the further away they are situated on a chromosome, the higher the recombination frequency between two loci). Markers located on different chromosomes are unlinked. Markers located far apart on the same chromosome are behaving as unlinked, because of high probabilities of one or more recombination events occurring. In a diploid, unlinked loci have an expected recombination frequency of 50%.
Often, the physical distance (in terms of DNA base pairs) between loci is not known. Recombination frequencies can be calculated in the offspring and be used to calculate the genetic distance between them.
The unit of genetic distance is the centi-Morgan (cM). A centi-Morgan map unit is defined as an expected cross-over frequency of 1 percent, or 0.01 per gamete formed. Generally, loci that have a recombination frequency of about 50% are considered 'unlinked'. However, they may belong to the same linkage group! The figure below shows a linkage map. This is a graphical representation of all the genetic positions of markers and genes relative to each other. Loci A and J at the far ends of the same chromosome behave as being not linked (recombination about 50%), since there will be a high frequency of one or more recombinations between them. They are, however linked via the loci B, C, ... and I.
![]()
Mapping functions are used to convert recombination frequencies into cM map units. The relationship between map distance and recombination frequency is non-linear: when the distance increases, the recombination tends to 50%.
Recombination frequencies are non-additive due to the possibility of even numbers of recombination events which are not observed from the genotype frequencies. Map distances in cM units are additive. So, if a locus B is located between loci A and C, the distance in centiMorgan units between A and B and between B and C can be added up to give the distance between A and C. In contrast, the recombination frequency between A and C is smaller than the summed recombination frequencies of A-B and B-C due to the possibility of double recombination events: a recombination in both A-B and B-C does not lead to (observed) recombination between A and C, it would be observed as a non-recombinant rather than two consecutive recombination events. A recombination frequency estimate from an observed number of recombinant genotypes will therefore, especially for loci linked at larger distances, often be an underestimation of the true number of recombination events, crossovers.
Summary
→ A recombination frequency indicates genetic distances between a pair of genes, a pair of markers or a gene and a marker
→ The closer two loci are situated to each other on a chromosome, the smaller the recombination frequency, and vice versa
→ The farther apart two loci are on a chromosome, the closer to 50% the recombination frequency will be. Loci on different chromosomes have an expected recombination frequency of 50% as well, corresponding to independent segregation at the two loci.
→ Genetic distances are usually indicated in centiMorgan units rather than recombination frequency units since cM distances are additive and recombination frequencies are not
→ The relationship between recombination frequency and cM genetic distance is non-linear
I want an alternative explanation | Move on to: Why use linkage maps?