SNPs
|
|
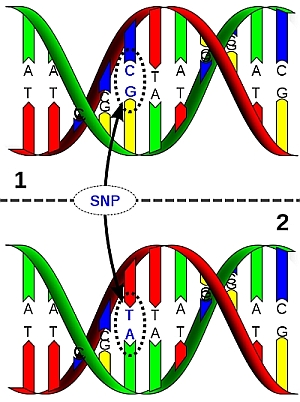
DNA sequences of homologous chromosome regions can be determined and compared between members of the population of the species. The sequence is often conserved: that means that the base sequence is almost identical between the individuals. This is in particularly true for genes (coding regions) and less so for non-coding regions. Comparison of conserved base sequences between individuals may reveal that they differ for only one base pair (nucleotide) in a stretch of DNA. This is called a Single Nucleotide Polymorphism (SNP, pronounce as "snip"). Such a polymorphism can be visualized as follows (figure below): During sequencing, the nucleotide sequence of a stretch of DNA is determined. Depending on the sequencing technique used (not treated here), this is observed as peaks of different colors (see example in figure), bars, or spots, representing the nucleotides in the DNA. The whole process is highly automated, e.g. peaks are read by special cameras and analyzed directly by a computer. SNP markers are often based on coding regions, since those are relatively conserved. SNPs that occur in the gene of interest itself (instead of on some gene that happens to be linked to the gene of interest), are the ideal marker to be used in marker-assisted selection. The association between the SNP allele and the allele of interest cannot be disturbed by a rare recombination event (although recombination within a gene is not impossible, but very unlikely). When SNPs occur in a gene, they do not always alter the function of the gene, since different codons may encode the same aminoacid. A change in base pair that does NOT alter the aminoacid that is coded for is called a 'synonymous mutation'. A base pair change that leads to a different amino acid is called a 'non-synonymous mutation'. Not all aminoacid changes change the properties and function of a protein. So, many DNA variations are phenotypically neutral.
Both alleles can be made visible, so SNPs are codominant markers. |
|
DNA molecule 1 differs from DNA molecule 2 at a single base-pair location (a C/T polymorphism). |
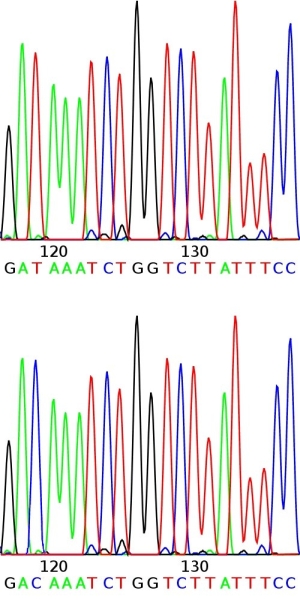 High-throughput SNP detection using sequencing techniques.
High-throughput SNP detection using sequencing techniques.
After identification of SNPs between the parents, the breeder may run many individuals in the progeny for such a SNP of interest. Such work is usually outsourced to specialized companies. The high-throughput running of SNPs needs knowledge of the DNA sequences flanking the SNP (that should not be polymorphic) and the bases that constitute the SNP. In this application we may run thousands of individuals for a certain SNP at relatively low cost.
There are also SNP platforms (spotted on chips, arrays), in which thousands of different SNPs may be run on individual DNA samples. In that case relatively few DNA samples are run against so many SNPs that a genome covering data set is obtained (about 100 € per DNA sample).For barley, for example, a set of 9000 SNP markers is available (the '9k Illumina Infinium iSELECT chip'). For each pair of barley cultivars, 2500 - 3000 of these SNPs are typically polymorphic.
Show/hide comprehension question...
Summary
→ SNPs are codominant markers
→ They occur relatively often, sometimes even within functional genes
→ SNPs are mostly detected by sequencing techniques
→ For a given SNP many plants can be screened efficiently by high-throughput techniques.