What is a genetic marker?
Chromosomes are very long (double) strands of DNA bases (DNA nucleotides). The bases of one strand correspond with counterpart bases on the other strand, and are called "base-pairs", often abbreviated as bp. The identity of the linearly arranged base pairs is called the DNA sequence. The base pairs together make a "story", based on an alphabet of only 4 letters (A, C, G, and T). A relatively small proportion of the chromosome consists of sequences that are expressed, and therefore contribute to the life function and appearance of the organism. The function of a large proportion of the DNA (formerly indicated as "junk DNA") is not yet fully understood. However, the sequences that are not expressed may still contribute to the function and appearance of the organism in some way. Within each species, and even between species, a lot of the DNA sequences are conserved (nearly identical), but also a lot is variable. The level of conservation is often sufficient to know which chromosome in one individual is equivalent (homologous) to which chromosome of another individual. However, between such homologous chromosome pairs, more or less variation in DNA sequence can be found. At the place (technically called "locus") of the gene for flower color, one chromosome may carry the allele for red flowers, the other for white flowers. The alleles at this locus hence will have a (somewhat) different DNA sequence. Between species there may also be larger structural rearrangements of (parts of) chromosomes. A whole set of tools has been developed to visualize variation in DNA sequence between homologous chromosome regions. A visualized difference in DNA sequence can be used as a "marker", to indicate ("mark") that region of the DNA strand. In principle for the whole genome (all DNA of the nucleus together) markers may be developed, both inside and outside genes.
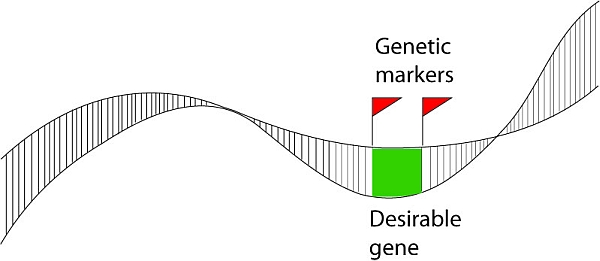
If a certain marker happens to be located near a gene for a useful trait, this marker may be of interest to the breeder. In case the gene that is near the marker occurs as one (favorable, for example resistance) allele on the chromosome of one parent (homozygous for that allele) and as a different (less favorable, for example susceptible) allele on the chromosome of the other parent, the marker (= visualized difference of a nearby DNA sequence) may be used to know which of the segregating progeny of a cross of these two parents has the one allele of the gene, and which the other, and hence which of the progeny to select. Also when the association between marker and gene is high (but maybe not exactly 100%), it may still be useful as a fast and cheap way to screen for resistance without the need of phenotyping the resistance of the plants.
Recombination
The distance between the sequence on which the marker is based and the gene of interest is relevant. The larger the distance between the marker locus and the locus of the gene of interest, the larger the chance that the chromosomes during meiosis perform a "crossover" between the loci. When the plant is heterozygous for both the marker and the gene, the gamete in which such a cross-over occurred in between the two loci, will show a "recombination": the marker allele that was linked to the favorable allele of the gene gets connected with the marker that indicated the unfavorable allele, or the marker that was linked to the unfavorable allele gets associated with the marker that indicated the favorable allele. The larger the distance between the marker locus and the locus of interest, the less reliable the association between the marker and the trait gene in inheritance is. This is because of the increased probability of recombination between the marker locus and the gene.
Summary
→ Genetic markers are useful for MAS when they are linked to genes of interest
→ The larger the distance between the marker locus and the locus of interest, the higher the chance of recombinations and thus the less reliable the association between the marker and the trait gene is