DNA markers
Historically, several types of DNA markers have been developed, introduced and applied. Most markers needed DNA amplication by PCR and separation on a gel. Methods to detect and visualize markers differed, leading to marker types as RAPD, RFLP, AFLP, SSR, CAPS and SNP markers. All of them had their advantages and disadvantages. Aspects considered to value a marker type include:
- reproducibility between labs and/or evaluations at the same lab
- a priori knowledge needed to develop the marker (if needed, the investment to develop a marker is high)
- numbers of markers that can be scored per reaction (the higher the better)
- numbers of alleles per marker (the higher the better)
- dominance or co-dominance of the marker (see below)
- possibility to run the marker in high throughput
Since most markers have been replaced by SNP markers now, and the others became more or less obsolete, we will not explain the methodology of all the marker types. One genetic property, however, will be explained: dominance/co-dominance of markers. This is a very important aspect to take into account when a breeder applies molecular markers.
Dominant versus codominant
When DNA markers of plant accessions are polymorphic, the polymorphism can be observed in two ways:
- In one parent, some marker allele is found (e.g. visible as one band on a gel) and in the other parent no marker allele is found (no band – figure below, left)
- In each parent a marker allele is found, e.g. visible as fragments with different lengths. On a gel this will be visible as two bands (figure below, right).
In situation 1, (presence or absence of an amplification product) presence of the amplification product is caused by an allele often indicated as '1'; absence by allele '0',(null-allele). The diploid homozygote 11 results in a band; the homozygote 00 results in no band; the heterozygote 10 results in an amplification product (and a band) which is identical to the product formed by parent 11 (figure below, left). Inheritance resembles that of a dominant trait. Therefore, it is called a dominant marker.
An essential aspect of dominance is that the heterozygote cannot be distinguished from one of the homozygoes, but that the other homozygote can still be distinguished from these two. In the case of codominance the heterozygote and the two homozygotes can all be distinguished from each other.
In situation 2, two types of bands are formed by fragments of different length: Parent 1 is homozygous and has AA alleles (alleles for long fragment) and parent 2 has aa (alleles for short fragment). The heterozygous offspring is Aa, so it has the alleles for both the long and for the short fragment, resulting in two bands on the gel.
In this case, the heterozygote and both homozygotes can all be distinguished from each other. This marker is called a codominant marker (Figure below, right).
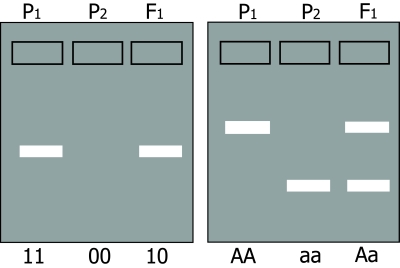
Comparison between dominant (left) and codominant (right) markers. Codominant markers can clearly discriminate between homozygotes and heterozygotes, whereas dominant markers do not. Genotypes at two marker loci (1 and A) are indicated below the gel diagrams.
For the dominant marker, locus 1 may carry two 1-alleles (1-1, or 11), two 0-alleles (0-0, or 00), or - in the heterozygote - one 1-allele and one 0-allele (1-0, or 10). In this example, the 0-allele results in absence of a band, for example caused by absence of an amplification product. Allele 1 results in presence of an amplification product and thus a band, regardless of the number of 1-alleles (one or two). Therefore, the heterozygote 10 and the homozygote 11 cannot be distinguished. For the co-dominant marker, allele A and allele a each result in a product, which is of different size, so the heterozygote can be distinguished from each of the homozygotes.
Some techniques that visualize variation in DNA sequences result predominantly in dominant markers; others in co-dominant markers. That depends on the strategy that is followed and on the nature of the sequence variation between the parents.
|
Dominant alleles of trait genes versus dominant alleles of markers In Mendelian genetics, dominant alleles of genes that code for traits overrule the effect of recessive alleles present. For example: if B is the dominant allele for hairy leaves and b is the recessive allele for smooth leaves, both BB and Bb plants have hairy leaves and bb plants have smooth leaves. In dominant markers, only dominant alleles can be detected as a band. So, on the gel, we see either a band or no band. Alleles that result in absence of a band are null-alleles: no band is formed, because the DNA fragment did not amplify. This could be caused by a number of different reasons. Therefore, in different individuals, different null-alleles might be present. Dominant markers are then sometimes coded as B_ versus bb). SNP markers are read in terms of the base pair that is involved: A, T, G or C. In the example in the picture on SNPs, the parents may be T, and G, respectively. SNP markers are co-dominant: if one chromosome carries the base pair AT and the other GC, (heterozygous), the marker is written as T/C. Note that also in genes that influence traits more than two alleles are often present. DNA-sequencing of genes has shown that in by far most genes, DNA polymorphism exist at population level. Some of those DNA differences may lead to a different trait expression, others are phenotypically neutral. Each of the DNA- sequence types may be considered a different allele. Also note that in the F1 of a cross of a cross pollinated species, e.g. apple, where both parents may be heterozygous, possibly for different alleles, more than two alleles (of a marker or a gene) may be segregating (B1B2 x B3B4) and there are different heterozygotes in the progeny that may or may not be distinguishable. In practical use, SNPs are biallelic, they allow just two variants to be visualized. |
Summary
→ Polymorphic markers indicate genotypic differences between the individuals tested.
→ It depends on the population tested whether a marker is monomorphic or polymorphic.
→ Co-dominant markers can discriminate between homozygotes and heterozygotes.
→ Dominant markers visualize only the presence/absence of a single marker allele and therefore do not allow a heterozygote for that allele to be distinguished from the homozygote for that allele.
→ The absent band in dominant markers represents a null-allele.
I want an alternative explanation | Move on to: Types of DNA markers