Alternative explanation: DNA markers: polymorphic or monomorphic; dominant or codominant
If a marker in a population has only one form, it cannot be used for selection. It is called a monomorphic marker. A polymorphic genetic marker can be useful for selection, when it is linked to a gene for a desirable trait.
We will now show three forms of DNA markers:
- Monomorphic markers
- Polymorphic markers which are dominant
- Polymorphic markers which are codominant
Monomorphic markers
|
|
|
|
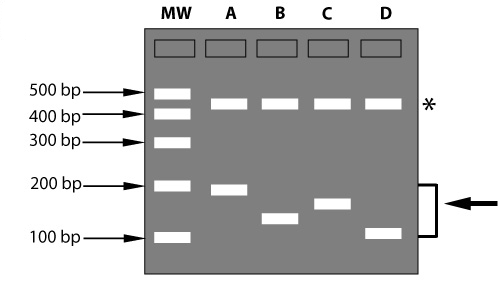
Top figure: if all tested individuals have the same allele, there is no polymorphism. The marker is a monomorphic marker (450 bp bands indicated with an asterisk). It does not reveal differences in alleles, and cannot be used for genetics or selection in breeding. Lower figure: allele 1 is present in all individuals tested. The part of the DNA between the restriction sites (red) can be made visible on a gel, using a so-called probe: a labeled DNA fragment that binds to specific DNA sequences and in that way 'stains' the DNA fragment.
Polymorphic markers which are dominant
|
|
|
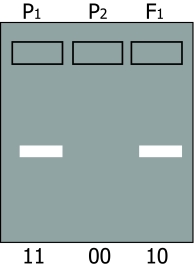
In a dominant polymorphic marker only one allele can be made visible. Let us assume that the procedure to make the marker visible involves:
- Cutting the DNA at the restriction sites (red) using restriction enzymes
- Amplifying the desired piece of DNA (dark blue) by PCR, resulting in numerous copies of this fragment
- Annealing a labeled probe to a certain part of the amplified product to make the band visible
Allele 1 results in a visible band on the gel. In allele 0 one restriction site is missing. The next restriction site could be too far away, resulting in a fragment that is too long to amplify. In case of a homozygote carrying allele 1 (11), or a heterozygote 10, a band will appear on the gel. In case of a homozygote carrying allele 0 (00), no band will appear on the gel.
Polymorphic markers which are codominant
|
|
|
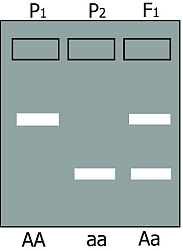
Codominant markers. For simplicity, we use here only two alleles: A and a (The gel on the right shows AA and aa parents and an Aa F1 An F2 population would give a segregation of all three patterns..
Codominant markers can discriminate between all possible gene combinations. Both alleles anneal to the same type of stained DNA probe, which is used to visualise the fragments as a band on a gel. The probe anneals to a specific region in the fragment. Outside the probe region, allele A has more sequence repeats than allele a. Because allele A is bigger than allele a, allele A will travel slower through an electrophoresis gel. A homozygote for allele A (so AA) will have one band (in fact two times the same (A+A) band). A homozygote for allele a (aa) will also have one band but this band will have moved faster (lower on the picture) than the band of allele A. Since a heterozygote (Aa) has a copy of allele A and a copy of allele a, a heterozygote will have both bands.